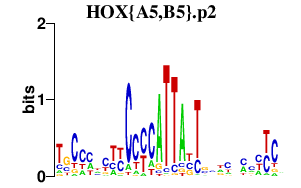
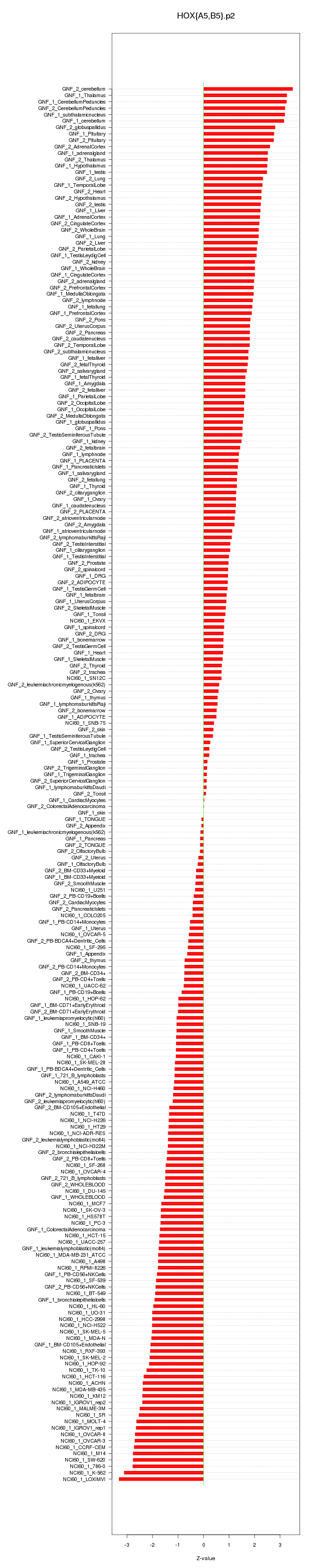
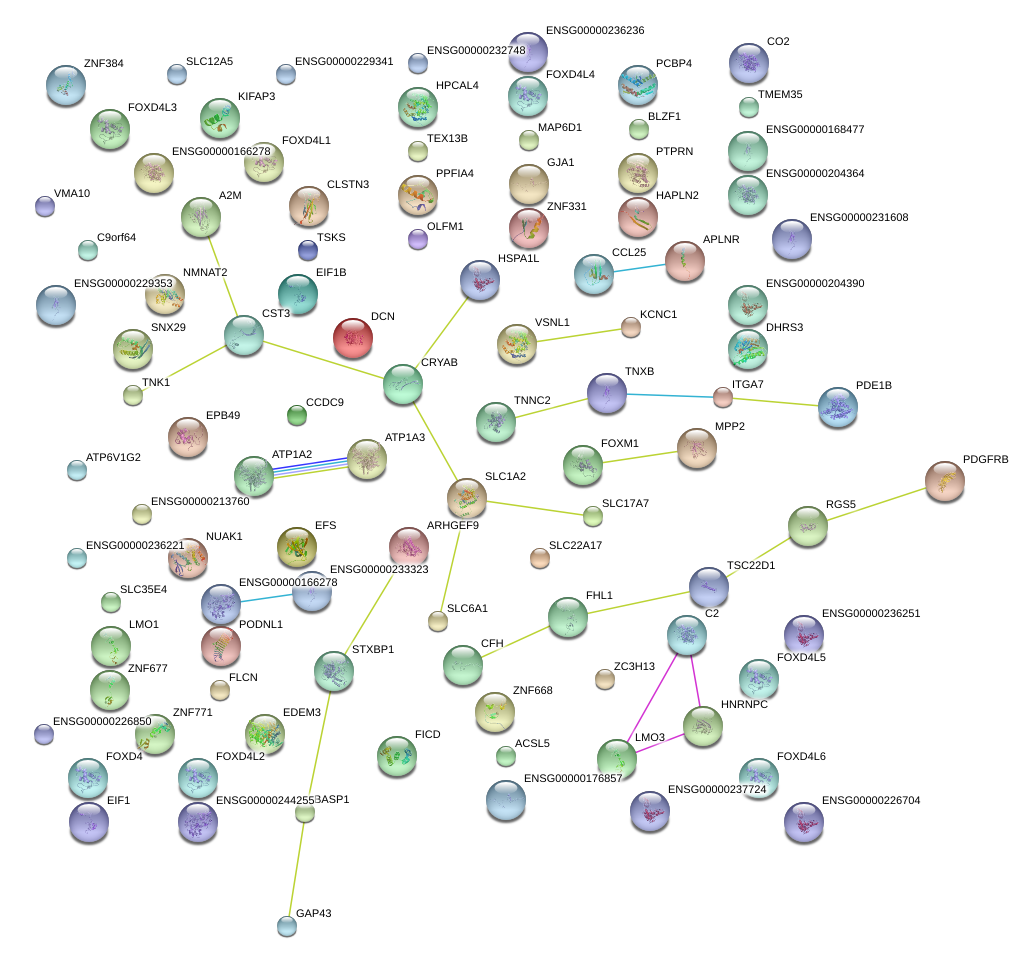
|
chr12_-_9268513
|
56.281
|
|
A2M
|
alpha-2-macroglobulin
|
|
chr2_+_17721806
|
44.208
|
NM_003385
|
VSNL1
|
visinin-like 1
|
|
chr12_-_6798419
|
39.566
|
|
ZNF384
|
zinc finger protein 384
|
|
chr20_+_44650260
|
36.780
|
NM_001134771
|
SLC12A5
|
solute carrier family 12 (potassium/chloride transporter), member 5
|
|
chr2_+_17721983
|
35.949
|
|
VSNL1
|
visinin-like 1
|
|
chr11_-_111782447
|
33.337
|
NM_001885
|
CRYAB
|
crystallin, alpha B
|
|
chr12_-_91572328
|
31.445
|
NM_133504
NM_133505
NM_133506
NM_133507
|
DCN
|
decorin
|
|
chr1_-_163113129
|
30.059
|
|
RGS5
|
regulator of G-protein signaling 5
|
|
chrM_+_12283
|
25.465
|
|
|
|
|
chr6_+_121756744
|
24.501
|
NM_000165
|
GJA1
|
gap junction protein, alpha 1, 43kDa
|
|
chr19_-_53757940
|
23.779
|
|
ZNF677
|
zinc finger protein 677
|
|
chr12_-_6798528
|
21.328
|
NM_133476
|
ZNF384
|
zinc finger protein 384
|
|
chr5_+_17217474
|
21.201
|
NM_006317
|
BASP1
|
brain abundant, membrane attached signal protein 1
|
|
chr3_+_115342302
|
20.998
|
|
GAP43
|
growth associated protein 43
|
|
chrX_+_135230736
|
20.987
|
NM_001159700
|
FHL1
|
four and a half LIM domains 1
|
|
chr1_-_183274008
|
20.723
|
NM_170706
|
NMNAT2
|
nicotinamide nucleotide adenylyltransferase 2
|
|
chr1_-_184723952
|
20.455
|
|
EDEM3
|
ER degradation enhancer, mannosidase alpha-like 3
|
|
chr11_-_62472901
|
19.570
|
|
|
|
|
chr13_-_45048436
|
19.088
|
NM_001243798
|
TSC22D1
|
TSC22 domain family, member 1
|
|
chr12_+_54955226
|
18.985
|
NM_001165975
|
PDE1B
|
phosphodiesterase 1B, calmodulin-dependent
|
|
chrX_+_100333835
|
18.009
|
NM_021637
|
TMEM35
|
transmembrane protein 35
|
|
chr6_-_31981049
|
17.925
|
NM_032470
|
TNXB
|
tenascin XB
|
|
chr12_-_56101465
|
17.842
|
|
ITGA7
|
integrin, alpha 7
|
|
chr9_-_86571652
|
17.071
|
NM_032307
|
C9orf64
|
chromosome 9 open reading frame 64
|
|
chr11_-_35440514
|
16.441
|
|
SLC1A2
|
solute carrier family 1 (glial high affinity glutamate transporter), member 2
|
|
chr1_-_170043608
|
16.224
|
NM_001204514
NM_001204516
NM_014970
|
KIFAP3
|
kinesin-associated protein 3
|
|
chr3_+_11034436
|
16.208
|
|
SLC6A1
|
solute carrier family 6 (neurotransmitter transporter, GABA), member 1
|
|
chr19_-_49944613
|
16.103
|
NM_020309
|
SLC17A7
|
solute carrier family 17 (sodium-dependent inorganic phosphate cotransporter), member 7
|
|
chr16_-_31085503
|
15.635
|
NM_024706
|
ZNF668
|
zinc finger protein 668
|
|
chr9_+_130374466
|
15.301
|
NM_001032221
NM_003165
|
STXBP1
|
syntaxin binding protein 1
|
|
chr8_+_21916858
|
14.927
|
|
EPB49
|
erythrocyte membrane protein band 4.9 (dematin)
|
|
chr14_-_23834417
|
14.864
|
|
EFS
|
embryonal Fyn-associated substrate
|
|
chr1_+_160085519
|
14.629
|
NM_000702
|
ATP1A2
|
ATPase, Na+/K+ transporting, alpha 2 polypeptide
|
|
chr2_-_220173693
|
14.574
|
NM_001199764
|
PTPRN
|
protein tyrosine phosphatase, receptor type, N
|
|
chr6_+_31868775
|
14.543
|
NM_001178063
|
C2
|
complement component 2
|
|
chrX_-_107225588
|
14.476
|
NM_031273
|
TEX13B
|
testis expressed 13B
|
|
chr5_-_149535138
|
14.423
|
|
PDGFRB
|
platelet-derived growth factor receptor, beta polypeptide
|
|
chr19_-_14064192
|
14.340
|
NM_001146254
NM_001146255
|
PODNL1
|
podocan-like 1
|
|
chr12_-_9268522
|
13.894
|
NM_000014
|
A2M
|
alpha-2-macroglobulin
|
|
chr1_-_12677347
|
13.811
|
NM_004753
|
DHRS3
|
dehydrogenase/reductase (SDR family) member 3
|
|
chr1_+_169337193
|
13.561
|
NM_003666
|
BLZF1
|
basic leucine zipper nuclear factor 1
|
|
chr3_-_183543295
|
13.391
|
NM_024871
|
MAP6D1
|
MAP6 domain containing 1
|
|
chr20_-_23618382
|
13.167
|
NM_000099
|
CST3
|
cystatin C
|
|
chr11_+_17757494
|
13.164
|
NM_001112741
NM_004976
|
KCNC1
|
potassium voltage-gated channel, Shaw-related subfamily, member 1
|
|
chr8_+_21916685
|
12.804
|
NM_001114138
NM_001978
|
EPB49
|
erythrocyte membrane protein band 4.9 (dematin)
|
|
chr19_+_8117887
|
12.742
|
|
CCL25
|
chemokine (C-C motif) ligand 25
|
|
chr19_-_50266514
|
12.710
|
NM_021733
|
TSKS
|
testis-specific serine kinase substrate
|
|
chr3_-_51993955
|
12.467
|
|
PCBP4
|
poly(rC) binding protein 4
|
|
chr12_-_9268475
|
12.384
|
|
A2M
|
alpha-2-macroglobulin
|
|
chr17_-_17140492
|
12.365
|
NM_144606
NM_144997
|
FLCN
|
folliculin
|
|
chr1_+_203020310
|
12.264
|
NM_015053
|
PPFIA4
|
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 4
|
|
chr3_+_11034419
|
12.139
|
NM_003042
|
SLC6A1
|
solute carrier family 6 (neurotransmitter transporter, GABA), member 1
|
|
chr1_+_156589085
|
12.112
|
NM_021817
|
HAPLN2
|
hyaluronan and proteoglycan link protein 2
|
|
chr22_+_31031672
|
11.999
|
NM_001001479
|
SLC35E4
|
solute carrier family 35, member E4
|
|
chr20_-_44455932
|
11.924
|
NM_003279
|
TNNC2
|
troponin C type 2 (fast)
|
|
chr11_-_57004674
|
11.856
|
NM_005161
|
APLNR
|
apelin receptor
|
|
chr19_+_47759802
|
11.719
|
|
CCDC9
|
coiled-coil domain containing 9
|
|
chr12_+_108909049
|
11.527
|
NM_007076
|
FICD
|
FIC domain containing
|
|
chr16_+_12070501
|
11.370
|
NM_032167
|
SNX29
|
sorting nexin 29
|
|
chr19_+_47759730
|
11.357
|
NM_015603
|
CCDC9
|
coiled-coil domain containing 9
|
|
chr19_+_54057898
|
11.209
|
NM_001253801
|
ZNF331
|
zinc finger protein 331
|
|
chr13_-_46626845
|
11.135
|
NM_015070
|
ZC3H13
|
zinc finger CCCH-type containing 13
|
|
chr17_+_39845142
|
11.043
|
|
EIF1
|
eukaryotic translation initiation factor 1
|
|
chr12_-_106533810
|
10.983
|
NM_014840
|
NUAK1
|
NUAK family, SNF1-like kinase, 1
|
|
chr12_+_7282963
|
10.966
|
NM_014718
|
CLSTN3
|
calsyntenin 3
|
|
chr12_-_16759939
|
10.859
|
NM_001243613
|
LMO3
|
LIM domain only 3 (rhombotin-like 2)
|
|
chrX_-_62975005
|
10.790
|
NM_001173480
NM_015185
|
ARHGEF9
|
Cdc42 guanine nucleotide exchange factor (GEF) 9
|
|
chr6_-_31514620
|
10.763
|
NM_001204078
NM_130463
NM_138282
|
ATP6V1G2
|
ATPase, H+ transporting, lysosomal 13kDa, V1 subunit G2
|
|
chr17_-_5404304
|
10.752
|
NM_001162371
|
LOC728392
|
uncharacterized LOC728392
|
|
chr9_+_42717233
|
10.731
|
NM_001099279
NM_199244
|
FOXD4L2
FOXD4L4
|
forkhead box D4-like 2
forkhead box D4-like 4
|
|
chr14_-_23822025
|
10.666
|
NM_016609
|
SLC22A17
|
solute carrier family 22, member 17
|
|
chr17_-_41984888
|
10.550
|
|
MPP2
|
membrane protein, palmitoylated 2 (MAGUK p55 subfamily member 2)
|
|
chr9_+_137967401
|
10.544
|
|
OLFM1
|
olfactomedin 1
|
|
chr2_+_113816214
|
10.243
|
NM_173170
|
IL36RN
|
interleukin 36 receptor antagonist
|
|
chr10_+_114136014
|
10.207
|
|
ACSL5
|
acyl-CoA synthetase long-chain family member 5
|
|
chr12_-_9268440
|
10.106
|
|
A2M
|
alpha-2-macroglobulin
|
|
chr16_+_30418885
|
10.086
|
|
ZNF771
|
zinc finger protein 771
|
|
chr6_-_31782786
|
10.075
|
NM_005527
|
HSPA1L
|
heat shock 70kDa protein 1-like
|
|
chr17_+_7284292
|
9.892
|
NM_001251902
NM_003985
|
TNK1
|
tyrosine kinase, non-receptor, 1
|
|
chr4_-_176733413
|
9.792
|
|
GPM6A
|
glycoprotein M6A
|
|
chr17_+_7623038
|
9.732
|
NM_020877
|
DNAH2
|
dynein, axonemal, heavy chain 2
|
|
chr19_+_7694658
|
9.693
|
NM_001171155
|
C19orf79
|
chromosome 19 open reading frame 79
|
|
chr8_-_143999255
|
9.680
|
NM_000498
|
CYP11B2
|
cytochrome P450, family 11, subfamily B, polypeptide 2
|
|
chr1_-_203320249
|
9.622
|
|
|
|
|
chr3_+_49027281
|
9.576
|
NM_177938
NM_177939
|
P4HTM
|
prolyl 4-hydroxylase, transmembrane (endoplasmic reticulum)
|
|
chr2_-_217540722
|
9.570
|
|
IGFBP5
|
insulin-like growth factor binding protein 5
|
|
chr13_-_46679143
|
9.542
|
NM_001872
NM_016413
|
CPB2
|
carboxypeptidase B2 (plasma)
|
|
chrX_-_62974987
|
9.509
|
|
ARHGEF9
|
Cdc42 guanine nucleotide exchange factor (GEF) 9
|
|
chr12_-_4488707
|
9.484
|
NM_020638
|
FGF23
|
fibroblast growth factor 23
|
|
chr19_-_17958814
|
9.422
|
NM_000215
|
JAK3
|
Janus kinase 3
|
|
chr6_-_29600959
|
9.374
|
NM_001470
NM_021904
|
GABBR1
|
gamma-aminobutyric acid (GABA) B receptor, 1
|
|
chr9_-_34710062
|
9.245
|
NM_002989
|
CCL21
|
chemokine (C-C motif) ligand 21
|
|
chr9_-_70429730
|
9.239
|
NM_001099279
NM_199244
|
FOXD4L2
FOXD4L4
|
forkhead box D4-like 2
forkhead box D4-like 4
|
|
chr1_+_203444955
|
9.216
|
|
PRELP
|
proline/arginine-rich end leucine-rich repeat protein
|
|
chr19_+_41515495
|
9.054
|
|
CYP2B6
|
cytochrome P450, family 2, subfamily B, polypeptide 6
|
|
chr12_+_7167979
|
8.937
|
NM_001734
NM_201442
|
C1S
|
complement component 1, s subcomponent
|
|
chr3_-_133648517
|
8.896
|
NM_025041
|
C3orf36
|
chromosome 3 open reading frame 36
|
|
chr3_-_127541160
|
8.891
|
NM_007283
|
MGLL
|
monoglyceride lipase
|
|
chr13_-_25496913
|
8.863
|
|
CENPJ
|
centromere protein J
|
|
chr13_+_58205943
|
8.824
|
|
PCDH17
|
protocadherin 17
|
|
chr3_+_39509069
|
8.766
|
NM_182935
|
MOBP
|
myelin-associated oligodendrocyte basic protein
|
|
chr1_+_4714668
|
8.689
|
|
AJAP1
|
adherens junctions associated protein 1
|
|
chr20_+_3024267
|
8.603
|
NM_001501
NM_178331
NM_178332
|
GNRH2
|
gonadotropin-releasing hormone 2
|
|
chr12_+_7013896
|
8.571
|
NM_001135217
NM_006992
NM_201650
|
LRRC23
|
leucine rich repeat containing 23
|
|
chr6_+_31082526
|
8.559
|
NM_014068
|
PSORS1C1
|
psoriasis susceptibility 1 candidate 1
|
|
chr17_-_41985112
|
8.541
|
NM_005374
|
MPP2
|
membrane protein, palmitoylated 2 (MAGUK p55 subfamily member 2)
|
|
chr3_-_42917632
|
8.526
|
NM_004391
|
CYP8B1
|
cytochrome P450, family 8, subfamily B, polypeptide 1
|
|
chr11_-_57283145
|
8.520
|
NM_003627
|
SLC43A1
|
solute carrier family 43, member 1
|
|
chr3_-_46506342
|
8.438
|
NM_002343
|
LTF
|
lactotransferrin
|
|
chr11_-_117102810
|
8.424
|
NM_004716
|
PCSK7
|
proprotein convertase subtilisin/kexin type 7
|
|
chr11_-_114271052
|
8.364
|
NM_019021
|
C11orf71
|
chromosome 11 open reading frame 71
|
|
chr14_-_23834378
|
8.345
|
|
EFS
|
embryonal Fyn-associated substrate
|
|
chr15_+_43809805
|
8.305
|
NM_002373
|
MAP1A
|
microtubule-associated protein 1A
|
|
chr16_-_49860917
|
8.240
|
NM_015069
|
ZNF423
|
zinc finger protein 423
|
|
chr22_-_27013959
|
8.212
|
NM_001887
|
CRYBB1
|
crystallin, beta B1
|
|
chr19_+_57999078
|
7.995
|
NM_001098491
NM_001098492
NM_001098493
NM_001098494
NM_001098495
NM_001098496
NM_024691
|
ZNF419
|
zinc finger protein 419
|
|
chr1_+_205236591
|
7.969
|
|
TMCC2
|
transmembrane and coiled-coil domain family 2
|
|
chr16_-_71610948
|
7.928
|
NM_000353
|
TAT
|
tyrosine aminotransferase
|
|
chr4_+_41258909
|
7.879
|
|
UCHL1
|
ubiquitin carboxyl-terminal esterase L1 (ubiquitin thiolesterase)
|
|
chr1_+_10490774
|
7.847
|
NM_001243768
|
APITD1-CORT
|
APITD1-CORT readthrough
|
|
chr1_+_114522029
|
7.780
|
NM_020190
|
OLFML3
|
olfactomedin-like 3
|
|
chr1_+_220701800
|
7.772
|
|
MARK1
|
MAP/microtubule affinity-regulating kinase 1
|
|
chr1_-_120935893
|
7.748
|
|
FCGR1B
|
Fc fragment of IgG, high affinity Ib, receptor (CD64)
|
|
chr1_-_23810627
|
7.743
|
|
ASAP3
|
ArfGAP with SH3 domain, ankyrin repeat and PH domain 3
|
|
chr13_-_25497054
|
7.735
|
NM_018451
|
CENPJ
|
centromere protein J
|
|
chr10_+_50817140
|
7.667
|
NM_020984
|
CHAT
|
choline O-acetyltransferase
|
|
chr16_+_30968614
|
7.585
|
NM_014712
|
SETD1A
|
SET domain containing 1A
|
|
chr5_+_149569519
|
7.576
|
NM_014228
|
SLC6A7
|
solute carrier family 6 (neurotransmitter transporter, L-proline), member 7
|
|
chr14_+_76618272
|
7.564
|
|
C14orf118
|
chromosome 14 open reading frame 118
|
|
chr16_+_86544104
|
7.507
|
NM_001451
|
FOXF1
|
forkhead box F1
|
|
chrX_+_8432870
|
7.477
|
NM_001001888
|
VCX3B
VCX3A
|
variable charge, X-linked 3B
variable charge, X-linked 3A
|
|
chr19_-_8642232
|
7.469
|
|
MYO1F
|
myosin IF
|
|
chr10_+_114135937
|
7.454
|
NM_016234
|
ACSL5
|
acyl-CoA synthetase long-chain family member 5
|
|
chr12_+_50497790
|
7.426
|
NM_005276
|
GPD1
|
glycerol-3-phosphate dehydrogenase 1 (soluble)
|
|
chr9_-_70178814
|
7.385
|
NM_001126334
|
FOXD4L5
|
forkhead box D4-like 5
|
|
chr9_+_137967088
|
7.331
|
NM_014279
NM_006334
|
OLFM1
|
olfactomedin 1
|
|
chr16_-_67427335
|
7.312
|
NM_015964
NM_016140
|
TPPP3
|
tubulin polymerization-promoting protein family member 3
|
|
chr4_-_114900802
|
7.276
|
NM_024590
|
ARSJ
|
arylsulfatase family, member J
|
|
chr9_+_137967366
|
7.260
|
|
OLFM1
|
olfactomedin 1
|
|
chr4_+_55523906
|
7.254
|
NM_000222
NM_001093772
|
KIT
|
v-kit Hardy-Zuckerman 4 feline sarcoma viral oncogene homolog
|
|
chr12_-_6798619
|
7.238
|
NM_001039920
NM_001135734
|
ZNF384
|
zinc finger protein 384
|
|
chrX_-_6453158
|
7.228
|
NM_016379
|
VCX2
VCX3A
|
variable charge, X-linked 2
variable charge, X-linked 3A
|
|
chr6_+_31588524
|
7.191
|
|
PRRC2A
|
proline-rich coiled-coil 2A
|
|
chr9_+_27109138
|
7.154
|
NM_000459
|
TEK
|
TEK tyrosine kinase, endothelial
|
|
chr17_-_8063946
|
7.135
|
|
VAMP2
|
vesicle-associated membrane protein 2 (synaptobrevin 2)
|
|
chr10_+_114135965
|
7.079
|
|
ACSL5
|
acyl-CoA synthetase long-chain family member 5
|
|
chr6_-_29600859
|
6.922
|
|
GABBR1
|
gamma-aminobutyric acid (GABA) B receptor, 1
|
|
chr11_-_125550674
|
6.868
|
NM_001612
NM_020069
NM_020107
NM_020108
|
ACRV1
|
acrosomal vesicle protein 1
|
|
chrY_-_1281526
|
6.835
|
NM_001012288
NM_022148
|
CRLF2
|
cytokine receptor-like factor 2
|
|
chr1_+_46806389
|
6.832
|
NM_199044
|
NSUN4
|
NOP2/Sun domain family, member 4
|
|
chr19_+_8274179
|
6.798
|
NM_024552
|
CERS4
|
ceramide synthase 4
|
|
chr9_-_4741798
|
6.752
|
NM_001199855
|
AK3
|
adenylate kinase 3
|
|
chr9_-_34620499
|
6.749
|
|
DCTN3
|
dynactin 3 (p22)
|
|
chr4_+_41258895
|
6.738
|
NM_004181
|
UCHL1
|
ubiquitin carboxyl-terminal esterase L1 (ubiquitin thiolesterase)
|
|
chr19_+_50529276
|
6.702
|
|
ZNF473
|
zinc finger protein 473
|
|
chr1_+_202317798
|
6.701
|
NM_001167857
NM_001167858
NM_002481
|
PPP1R12B
|
protein phosphatase 1, regulatory subunit 12B
|
|
chr9_-_135819995
|
6.597
|
NM_000368
NM_001162426
NM_001162427
|
TSC1
|
tuberous sclerosis 1
|
|
chr3_-_164914448
|
6.408
|
NM_014926
|
SLITRK3
|
SLIT and NTRK-like family, member 3
|
|
chr16_-_89556886
|
6.398
|
NM_013275
|
ANKRD11
|
ankyrin repeat domain 11
|
|
chrX_-_137949716
|
6.394
|
NM_001139502
|
FGF13
|
fibroblast growth factor 13
|
|
chr2_+_113816684
|
6.391
|
NM_012275
|
IL36RN
|
interleukin 36 receptor antagonist
|
|
chr4_-_76944585
|
6.364
|
NM_001565
|
CXCL10
|
chemokine (C-X-C motif) ligand 10
|
|
chr20_-_1638359
|
6.349
|
NM_001039508
NM_018556
NM_080816
|
SIRPG
|
signal-regulatory protein gamma
|
|
chr22_-_36556781
|
6.336
|
NM_145640
|
APOL3
|
apolipoprotein L, 3
|
|
chr14_+_76618289
|
6.303
|
|
C14orf118
|
chromosome 14 open reading frame 118
|
|
chr6_-_29595746
|
6.282
|
|
GABBR1
|
gamma-aminobutyric acid (GABA) B receptor, 1
|
|
chr1_-_184723996
|
6.275
|
|
EDEM3
|
ER degradation enhancer, mannosidase alpha-like 3
|
|
chr11_-_117102758
|
6.270
|
|
PCSK7
|
proprotein convertase subtilisin/kexin type 7
|
|
chr4_-_57687827
|
6.250
|
NM_021114
|
SPINK2
|
serine peptidase inhibitor, Kazal type 2 (acrosin-trypsin inhibitor)
|
|
chr14_+_76618244
|
6.233
|
NM_017926
NM_017972
|
C14orf118
|
chromosome 14 open reading frame 118
|
|
chr7_+_90225607
|
6.221
|
|
CDK14
|
cyclin-dependent kinase 14
|
|
chr3_-_109056388
|
6.183
|
NM_018189
|
DPPA4
|
developmental pluripotency associated 4
|
|
chr10_+_115310589
|
6.155
|
NM_001177660
|
HABP2
|
hyaluronan binding protein 2
|
|
chr5_+_148520985
|
6.151
|
NM_014945
|
ABLIM3
|
actin binding LIM protein family, member 3
|
|
chr17_+_20771745
|
6.146
|
|
|
|
|
chr3_-_14989399
|
6.134
|
|
LOC100505641
|
uncharacterized LOC100505641
|
|
chr6_-_46922533
|
6.132
|
NM_015234
|
GPR116
|
G protein-coupled receptor 116
|
|
chr9_-_4742042
|
6.127
|
NM_001199853
|
AK3
|
adenylate kinase 3
|
|
chr16_-_2390711
|
6.093
|
|
ABCA3
|
ATP-binding cassette, sub-family A (ABC1), member 3
|
|
chr13_+_58205788
|
6.091
|
NM_001040429
|
PCDH17
|
protocadherin 17
|
|
chrX_+_57618440
|
6.086
|
|
ZXDB
|
zinc finger, X-linked, duplicated B
|
|
chr11_+_74862031
|
6.082
|
NM_001145212
NM_007256
|
SLCO2B1
|
solute carrier organic anion transporter family, member 2B1
|
|
chrX_-_104465349
|
6.036
|
NM_031274
|
TEX13A
|
testis expressed 13A
|
|
chr1_-_203154304
|
6.035
|
|
CHI3L1
|
chitinase 3-like 1 (cartilage glycoprotein-39)
|
|
chr7_+_156435670
|
6.034
|
NM_001184997
|
RNF32
|
ring finger protein 32
|
|
chrX_-_130423387
|
6.024
|
NM_001170961
NM_001170962
NM_001555
NM_205833
|
IGSF1
|
immunoglobulin superfamily, member 1
|
|
chr4_+_41258941
|
6.008
|
|
UCHL1
|
ubiquitin carboxyl-terminal esterase L1 (ubiquitin thiolesterase)
|
|
chr15_-_26108869
|
5.942
|
|
ATP10A
|
ATPase, class V, type 10A
|
|
chr1_+_203734283
|
5.926
|
NM_001136190
NM_017773
|
LAX1
|
lymphocyte transmembrane adaptor 1
|
|
chr11_+_62475165
|
5.852
|
|
GNG3
|
guanine nucleotide binding protein (G protein), gamma 3
|
|
chr16_+_57769626
|
5.793
|
NM_005886
|
KATNB1
|
katanin p80 (WD repeat containing) subunit B 1
|
|
chr4_-_48014960
|
5.777
|
NM_000087
|
CNGA1
|
cyclic nucleotide gated channel alpha 1
|
|
chr8_-_66701175
|
5.774
|
NM_002603
|
PDE7A
|
phosphodiesterase 7A
|
|
chr20_-_56100138
|
5.750
|
NM_080618
|
CTCFL
|
CCCTC-binding factor (zinc finger protein)-like
|
|
chr5_+_161274939
|
5.711
|
NM_001127644
|
GABRA1
|
gamma-aminobutyric acid (GABA) A receptor, alpha 1
|
|
chr8_+_26371461
|
5.686
|
NM_001197293
|
DPYSL2
|
dihydropyrimidinase-like 2
|
|
chr3_-_109056333
|
5.684
|
|
DPPA4
|
developmental pluripotency associated 4
|
|
chr16_-_58005011
|
5.605
|
NM_001135639
NM_001297
|
CNGB1
|
cyclic nucleotide gated channel beta 1
|
|
chr12_-_8218930
|
5.561
|
NM_004054
|
C3AR1
|
complement component 3a receptor 1
|
|
chr6_+_31554975
|
5.523
|
NM_001166538
NM_007161
NM_205840
|
LST1
|
leukocyte specific transcript 1
|